Image Processing for Tumor Detection
Computer Vision (CV)-based image processing pipeline for CT and Ultrasound imaging of suspected cancerous growths.
I developed this project as part of my Stanford undergraduate coursework in Bioengineering 103: Systems Physiology and Design.
PART 1: For the first half of the project, I analyzed lung CT scans. CT scans are more likely to show lung abnormalities, including infections and tumors, than routine chest X-rays. This is partially due to the fact that we can use CT scans to create detailed 3D reconstructions of the lungs composed from multiple cross-sectional slices, allowing for comprehensive visualization.
I started by taking in a set of lung CT scans and applying K-means clustering from Scikit-learn to segment the images by tissue type:
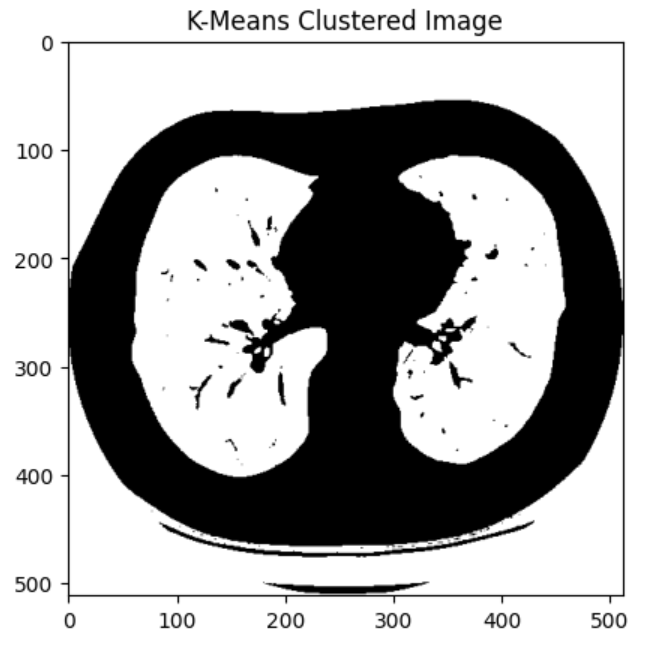
Then, I utilized OpenCV image processing functions to further isolate the lung tissue in the image segmentation:
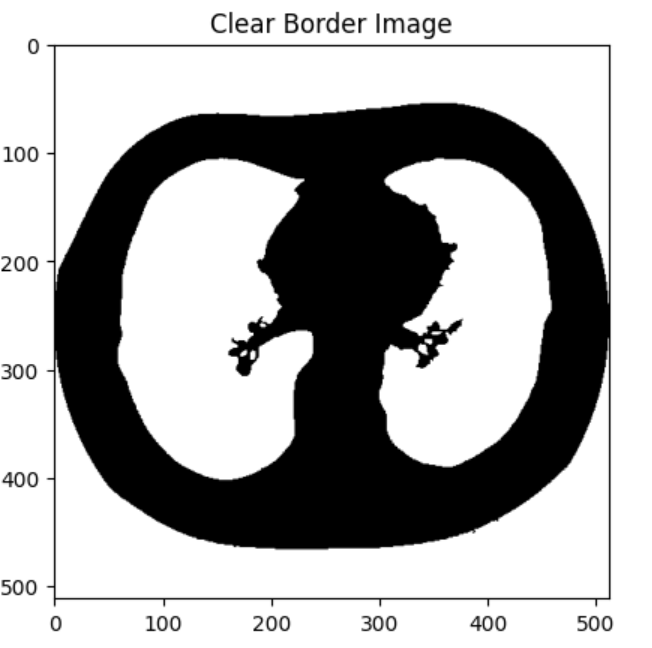


Finally, I applied these image masks to all of the CT scans in the collection, and was able to make a 3D reconstruction of the lungs:
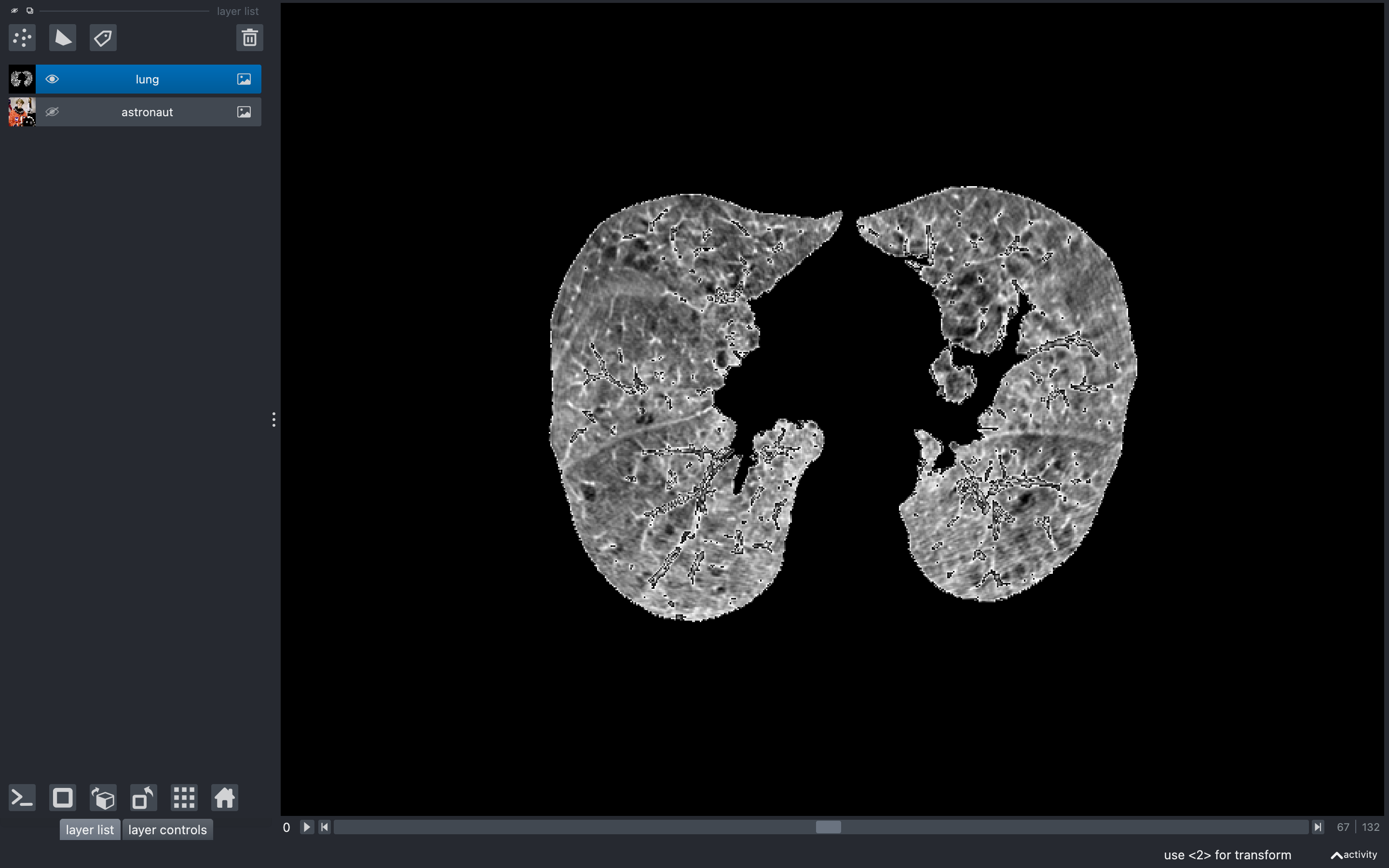
PART 2:
For the second half of the project, I shifted from CT to breast ultrasound*and explored how deep learning can support early breast cancer detection. Ultrasound is increasingly used as a rapid, point-of-care diagnostic tool, and in some cases can reveal early-stage cancers that are not detected by mammography.
Dataset
I worked with a breast ultrasound dataset labeled by oncology physicians with three classes: normal, benign*, and malignant. The dataset contains 780 PNG images (average size ~500×500) from 600 patients(ages 25–75).
Approach: tumor segmentation with Attention U-Net
I implemented an attention-based U-Net, which encodes the image into a compact representation and decodes it back into pixel-wise masks.
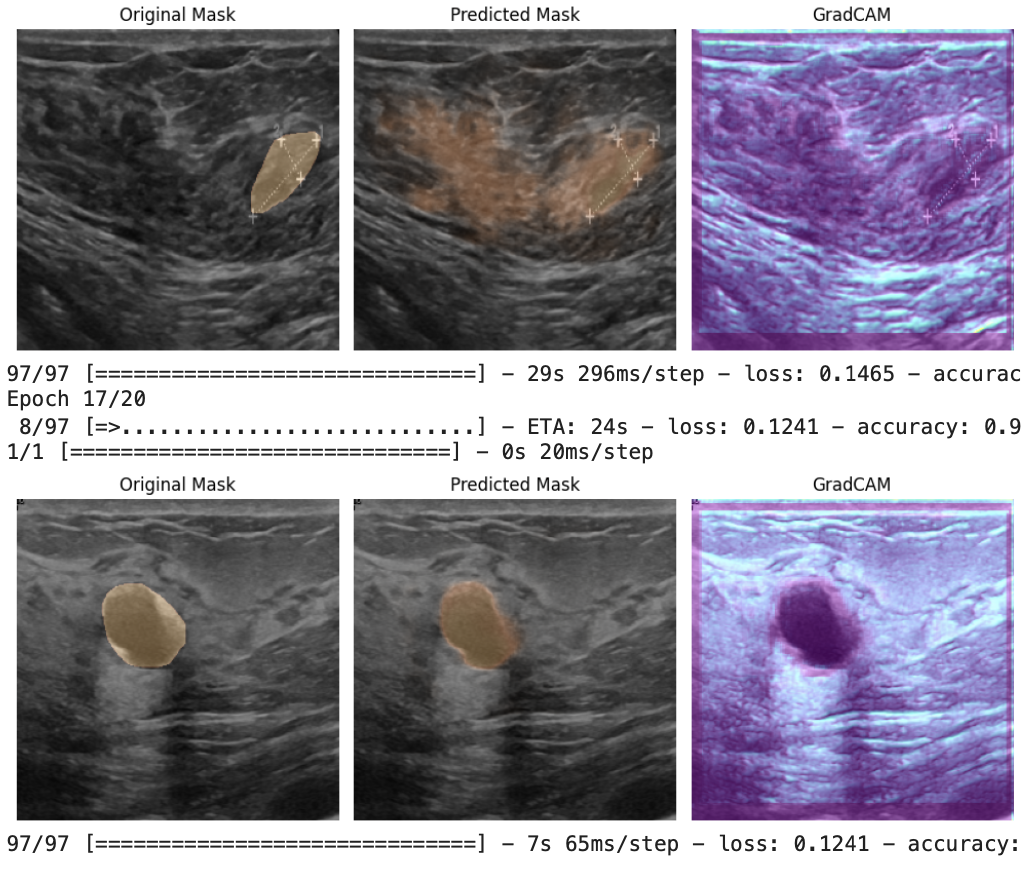
U-Net’s skip connections preserve fine spatial detail, and the attention mechanism helps the decoder focus on the most relevant regions (in this case, subtle tumor boundaries and context within heterogeneous ultrasound texture). This combination improved mask quality compared to a generic encoder–decoder model.
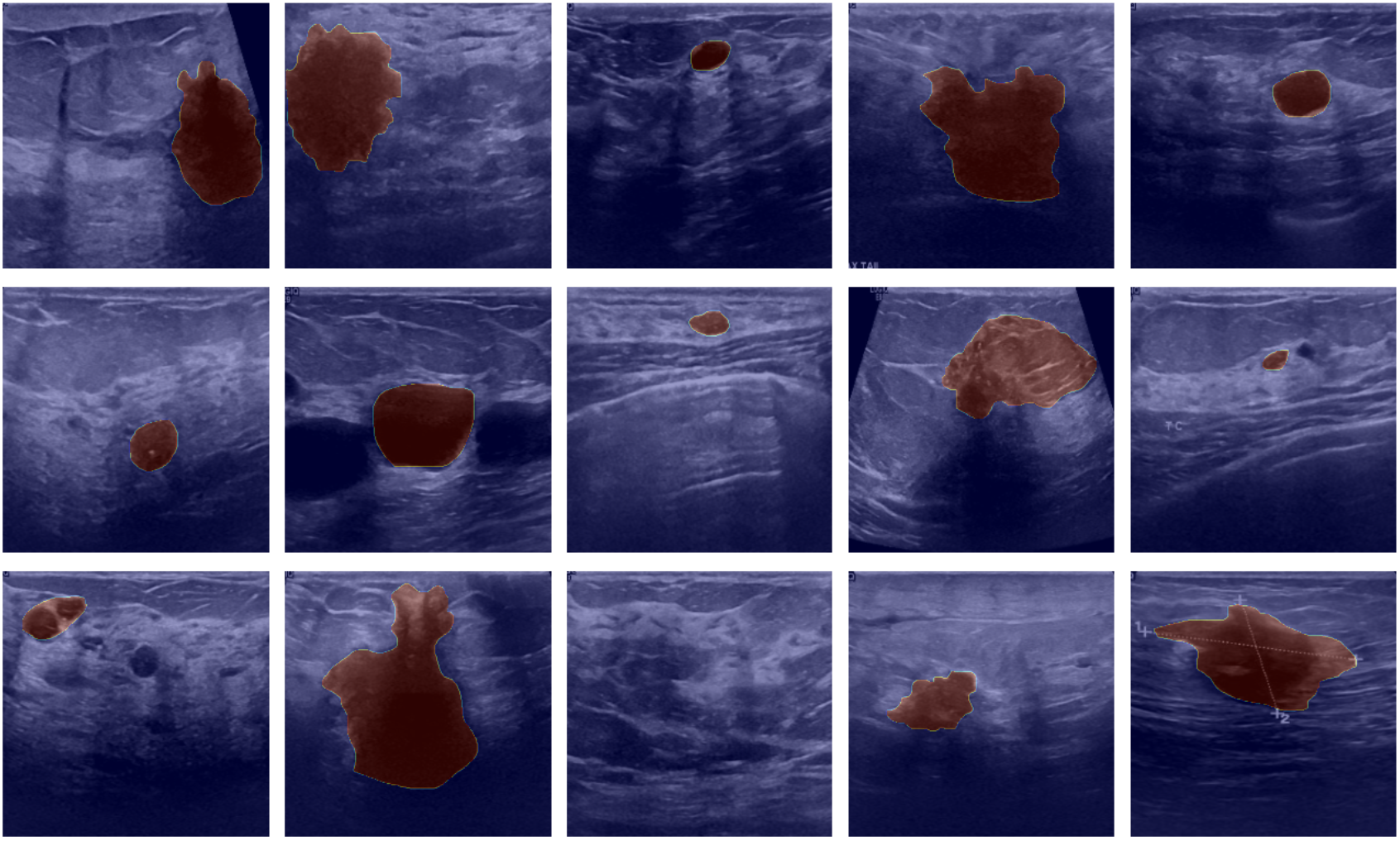
After training, I compared ground-truth masks to predictions and used Grad-CAM to visualize which regions most influenced the model’s segmentation decisions: